Actionable digital phenotyping: a framework for the delivery of just-in-time and longitudinal interventions in clinical healthcare
Introduction
The potential of mobile health is fueled by digital data captured in real time and real-world environments that allows quantification of the personal experience of illness. The myriad of sensors on today’s smartphones and smartwatches can help patients track exercise, sleep, heart rate, and much more. The concept of digital phenotyping, defined as the “moment-by-moment quantification of the individual-level human phenotype in situ using data from personal digital devices” (1) has emerged as a means to conceptualize the possibilities of such data to improve health and has been extensively applied in mental health (2). Yet the vast quantities of this new data have not yet transformed routine clinical care and still remain more a research topic than clinical tool to support prevention, clinical decision support, and personalized medicine. While there are numerous reasons for this discrepancy between data and utility, there is a clear need for solutions that can harness the potential of transforming mobile data into health data.
Consider the clinical case of a patient using a smartphone app over three months, responding to surveys about mood and anxiety and capturing real time data on steps and sleep. An intelligent machine learning algorithm used in the app is able to capture the baseline of this patient using a combination of sensors and surveys. This algorithm is then able to perform real time assessment of risk of relapse based on changes in these digital signals, notifying the patients, alerting the clinical team, and recommending digital interventions in real time. This case, as well as many others such as deploying emergency responses to a signal suggesting high risk of suicide based on geolocation, survey responses, and social activity, are possible today with advancements in technology and access to care. But little has been done to standardize the use and development of systems that support cases such as these in the modern digital clinic (3).
Adapting to ever-changing legislative, security, privacy, and medical needs, the Health Level Seven (HL7) International organization proposed the Fast Healthcare Interoperability Resources (FHIR) standard (4) to help make digital data more uniform and interpretable. FHIR aims to facilitate secure interoperation between medical records systems as well as third-party applications such as medical devices and smartphone apps. However, while FHIR proposes an easily generalizable and interoperable standard for medical records and devices, its capability to support such personalized, highly contextual sensor-driven data is still evolving. It is possible to envision how FHIR standards could be augmented and expanded to one day support such clinically useful applications (5).
As we approached the concept of real-time digital clinical interventions, we identified numerous apps capable of delivering health-related intervention but found in line with prior research that they required manual configuration and deployment with no feedback system in place for either the patient or clinician. Furthermore, many of the apps that others and our team identified lacked integration with other existing systems such as electronic medical records, a concern that could easily be mitigated through adoption of FHIR. Recognizing this need, we blueprinted new techniques to react to potentially rapid changes in mental health state such as threshold-triggered auto-deployment of interventions, feedback monitoring for advanced clinical decision support, and auto-configuration based on incoming patient data. We explored existing methods of data storage, encryption and access, future proofing, and interoperability in the context of mobile device based digital phenotyping and health interventions.
In this paper, we frame the discussion around a software protocol and smartphone platform to provide concrete examples of clinical use cases. The software protocol, called an Application Programming Interface in software engineering parlance, defines all properties of communication between parties. An ideal protocol should be easy to use but hard to misuse, self-documenting, flexible and extensible, yet concise (6). It can be thought of as a dictionary containing the core set of nouns and verbs as well as the spoken dialect itself. Together, a system, its protocol, and an ecosystem of third-party clients that speak this protocol all constitute a platform. We incrementally developed our platform by following industry standards validated alongside our core goals: data modeling (consisting of a well-designed object hierarchy, interaction patterns, storage mechanisms and exchange or transfer), extensibility, and interoperability. Through various stages of research and development, clinicians and patients were consulted and interviewed about their experiences, usage patterns, and criticism. These feedback then informed modifications to our architecture and implementation.
Discussion
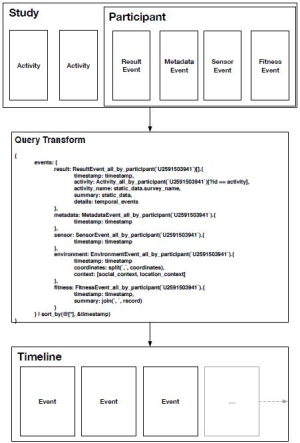
To organize the discussion, we present a theoretical framework, shown below in Figure 1. This paper aims to translate the clinical needs presented to the left of the pyramid into the engineering requirements on the right.
Data modeling
Resource schema
For data modeling, digital mobile health platforms need to ensure simplicity and efficiency, along with reliance on existing standards. We wanted to develop a protocol composed of smaller interconnected modular components. We began by defining the ‘schema’, a rigid structural vocabulary, with which data could be organized and interpreted, agreed upon in advance by all parties working with some data constrained by that schema. A succinct set of core data types in our schema, outlined in Table 1, is initially declared and further elaborated upon by subtypes with more specific use-cases and properties.

Full table
Ochian et al. (7) note that current challenges in healthcare informatics revolve around low-latency and high-throughput big data processing of both well-structured and un-structured but specialized medical data. The methods of data warehousing and integration are not novel to the biological or medical fields; Shah et al. (8) first demonstrated in 2005 a relational database approach to high-throughput data integration and analysis in bioinformatics. To facilitate such high frequency measurement recordings from on-device or external sensor instruments, we created special types of objects called ‘events’; as with droplets of water in a larger stream, events form a virtual ‘event stream’ that can be filtered and whose changes can be reacted upon in a dynamic manner.
To reduce complexity and promote ease of use of the protocol, we limited our vocabulary of specialized subtypes to only a few nouns, outlined in Table 2, that would be capable of build datasets that suit the changing needs of digital mental health research. FHIR on the other hand was designed to ensure compatibility across electronic medical records vendors and afford hospital systems the greatest amount of cross-talk, and so declares a vast set of specialized subtypes spanning over 50 nouns such as Patient, Condition, and Practitioner. For FHIR, this is a necessity that aims to organize data into highly complex structures capable of representing every capability, possibility, or outcome of a hospital system, including billing and insurance management. Though clinicians may find the set of types we have declared to be fairly limited in comparison to FHIR, we designed the protocol not to replace FHIR entirely but instead to enable its use as an adjunct where needed.

Full table
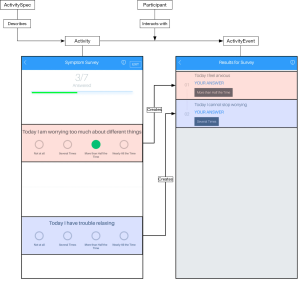
The concrete relationship of these virtual data types can be mapped spatially into the application’s user interface, as shown in Figure 2, but the protocol itself is able to be used in any kind of application, including those that do not present an interface at all. Another facet of the schema not immediately visualized, is reacting to events instead of repeatedly checking for them. The two event types, ActivityEvent and SensorEvent represent reactive data in event streams containing information contextually linked either to a Sensor equipped by a patient, or interaction with an Activity.
Interchange format
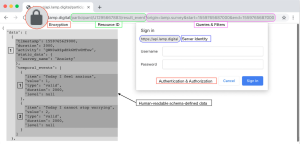
For digital health, it is important that clinical data is easily interpreted and visualized in any context. The JavaScript Object Notation (JSON) data interchange format, a well-accepted internet standard (9), was chosen because of its textually encoded representation of data in a human-readable and hierarchically organized way. Unlike some older formats such as the eXtensible Markup Language (XML), JSON does not require the explicit definition of schema. Though we have already defined a schema, avoiding ‘schema lock-in’ affords the protocol flexibility and extensibility, as explained in Section “Clinical intervention support”. Other formats were also briefly considered for their potentially superior encoding ability and minimal storage size, but ignoring the vast adoption of JSON across internet services, mobile apps, development frameworks and tools would have eliminated many opportunities for interoperability, again explored in Section “Clinical intervention support”. Furthermore, because most transmission and storage formats support compression, the textual nature of JSON could maintain human readability while also minimizing storage size, at the slight cost of computing time required for compression.
While FHIR does support JSON, this data is transformed post-hoc from the original XML document into inefficient and non-standard JSON documents. These documents, for example, represent keyword-indexed data linked to more data as an unsorted list of pairs. While seemingly trivial in notation, this minute structural modification forces the user’s computer to perform a time and processing-intensive unoptimized search through every element of an array instead of an optimized pre-calculated lookup operation; this would be as if one were to search for the definition of a word by reading every alphabetized definition preceding it, instead of jumping to the page containing the word’s first letter and scanning from there. Furthermore, these lists may now contain duplicate data that could go undetected where a dictionary would impose constraints on the uniqueness of data. By natively supporting JSON, the protocol is afforded considerable advantages in machine processing, reducing computing power, and time requirement. These advantages are then realized during computation tasks performed on cheaper mobile devices that must operate in conditions without stable internet connection to a more powerful server.
Transmission format
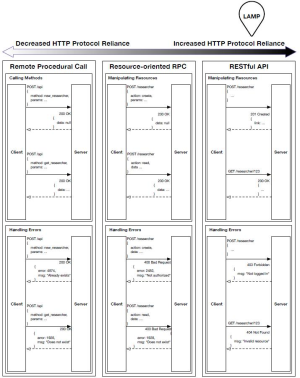
For digital health to support interoperability, protocols must describe how the systems that understand it are to speak to each other. The HyperText Transfer Protocol (HTTP) standard is one that the vast majority of internet services and mobile apps already rely on (including FHIR), and with much of the internet’s infrastructure already purpose-built for HTTP, important security and efficiency optimizations such as encrypting, compressing, and caching data as it moves from point to point are trivial. The Representational State Transfer (REST) model (10) describes interactions through a system of requests and responses by which an application can export ‘objects’ (also referred to as ‘documents’) over HTTP, each with a uniform resource locator (URL) that may be accessed as a link (11). A comparison against other approaches is detailed in Supplement I. As standard internet browsers support this HTTP and REST functionality, explained visually in Figure 3, they are able to present and manipulate data, though they may not semantically understand it. Any individual holding appropriate access rights may enter the URL corresponding to the data they wish to view and may interpret its decrypted contents using either a desktop or mobile browser.
Following standard guidelines, we supported the HTTP methods: GET to list entire collections of resources or read individual resources; POST to create resources; PUT and PATCH both to update resources; and DELETE to delete resources. With these verbs in place, actions are communicated from one entity to another in a format such as “GET/participant/
FHIR also uses the HTTP method OPTIONS to produce conformance resources talking about the object schema and capabilities of the server. FHIR servers do not actually delete resources when a deletion action is requested, but instead they are versioned, and these older versions can be retrieved from a “Recycle Bin” at a later time. We consider the OPTIONS method as well for use in polymorphic data (structured meta-information on the usage of some resource), service capabilities, or the transaction audit log. This audit log records modifications to data as well as all access requests for internal auditing and data recovery.
Documentation & discovery
Clinical systems require collaboration with diverse stakeholders. For maintainers, developers, and statisticians to easily explore, learn, and use the protocol, we document and make available the protocol using the robust and tightly integrated OpenAPI and JSON Schema meta-protocols. Integration with these meta-protocols enabled features we felt were vital to open-source platforms, such as the easy generation of client code, development tooling, and support for numerous other frameworks. For example, we were able to quickly generate a “Rosetta stone” for developers using many various programming languages to speak the protocol without handling any of the translation and communication. We chose to avoid schema documentation systems such as JSON-LD that required modifications to the data being presented from the server, adding complexity to the readability of data in JSON format, making it harder to read and debug. Another concern was the runtime requisition nature of JSON-LD, where the data itself is bundled with the schema, and requires recursively querying each element to build a tree of relationships between objects. OpenAPI instead declares in advance of even receiving a single piece of data, all relationships between elements as well as actions that may be taken on them.
Unlike FHIR and many other common systems where the protocol is static and expected to be unchanging, entities that speak the LAMP Protocol may maintain and provide information about their ‘dialect’ for other entities to reconfigure and reorient themselves. We also designed interaction responses to be self-evident by packaging the requested data in a document format with a “live citation,” consisting of the same access request details and security credentials as are saved to the aforementioned audit log. When examining data from a different time point, the audit log can be consulted with the “live citation” to produce the original data along with the set of all changes that have occurred since that point.
Network modeling
Document transformations
Supporting interoperability is critical for the success of any digital health platform. Some systems that use the protocol may need “glue code” that acts as a translator, reducing efficiency and wasting computing power. Several document manipulation frameworks able to be embedded within the protocol have evolved to both query and manipulate documents in an efficient manner; this functionality can be used in reverse to translate the LAMP Protocol into the FHIR standard for EMR integration. Boussadi et al. (12) and Wagholikar et al. (13) use a system-integrated approach to develop a FHIR layer above the i2b2 software, allowing FHIR queries to translate to the i2b2 database system. In contrast with their approach, FHIR integration with the LAMP Protocol is trivially implemented as a set of adapter queries using such a framework and does not require additional computing power or storage space for translating any functions of the data. The XML format used by FHIR supports a pair of frameworks called XPath and XQuery, outlined in Supplement I, to perform these tasks, but no such universally accepted framework exists for the JSON format. In response, HL7 developed the standards-incompatible “FHIRPath” framework to replicate these features in FHIR (14). The decision was made to support the “JSONPath” framework, used in industry by Amazon, Google, eBay, and others, though not ratified by any standards committee. With a concise implementation and large developer community contributing to implementations in many programming languages, the level of integration, support, performance, and quality of code would be higher than if we had developed a custom framework as HL7 did.
Tags
The expected lifespan of clinical systems suggests there is a need to support backwards compatibility, communication with other coexisting clinical systems, as well as future systems whose protocols may not exist today. Thus, we aimed to further develop integration with external data and future-proof the protocol. The protocol’s schema thus supports the “Tag” data type; though similar to the mechanism utilized by FHIR, XML DTD-based document extensions, the content of a Tag is lacking a schema. The protocol in this case declares that it does not understand or even directly handle the content of Tags, and instead acts only as a broker or middleman for two other entities that do understand the data to communicate. As they are out-of-line from the rest of the protocol, they may hold binary data, arbitrary strings, or well-structured objects. Though the protocol itself uses JSON Schema, within a Tag, the provision of schema, definition language, storage and access mechanism are left to other entities instead. A further benefit of Tags is the ad-hoc integration with clinical dashboards supporting patient, family, and care team messaging, as explained further in Section “Clinical intervention support”. Detail on the reference implementation is provided in Supplement II.
Automations
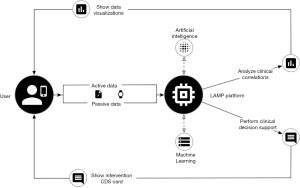
Digital health platforms must not only capture data but offer actionability towards improving patient health. Most REST-based protocols are designed without the ability to perform an action unlinked to a resource; that is, actions may only take the form of verbs attached to nouns, such as “CREATE activity_event”. FHIR supports “OperationDefinition,” the definition of a computable result yielded from an operation or complex query. Similarly, building upon Tags, we introduce Automations, resources that act as modular “applets” around static data, supporting different data formats and external resources, shown visually in Figure 4. Automations are run in isolated environments through carefully configured system containers, currently supporting the R, Python, and JavaScript languages. Though this incurs a slight latency cost in retrieving data from an Automation applet, it enables complete data and algorithmic reproducibility when used in tandem with “live citations.” Each container is identical to every other container of the same Automation, and so applets when run with some input identical at different time points yield the exact same output, also identical at the same time points. This reproducibility offers the key advantage of analytic validity within and between applet results, such as those producing visualizations that can be shown by another entity.
Containerized replication of programmable scientific methods has been shown before in bioinformatics by Challis et al. (15), Hung et al. (16), and Ríos et al. (17); we however present a layer of meta-programmability through the program code attached to Automations. By abstracting the semantics of system containers, package dependencies, configuration, and data pre-processing away from the user “installing” the Automation, we aim to reduce the level of systems engineering knowledge required to develop truly reproducible research methods. Where the ActivitySpec data type provides to the protocol reproducible patient-facing activities mirroring experimental methods, Automations mirror the role of post-collection data analysis linked to a specific context of data. Thus, all data collection, schema-validation, homogenization, and further post-processing steps are performed by the main LAMP Platform itself. Furthermore, these applets can be configured to automatically run when changes are detected on any Resource, or when Events enter the stream for a particular set of Participants, making the framework truly dynamic and researcher input or configuration no longer required.
Clinical intervention support
Clinical case
The protocol now forms the underpinnings of a simple, secure, and efficient pipeline for reproducible and actionable data and also builds native extensibility and interoperability with other entities and protocols. FHIR integration into existing EMR and billing systems would establish the ability to able to span the continuum of care in the hospital setting. Walking through the advanced features detailed in Section “Network modeling”, we share tangible use-cases from workflows currently in use in several research studies. Integrating custom functions into the document manipulation framework allows for building expressive data queries that stitch together different resources and events, aggregating calculation and computation, obtaining data from remote sources, and exporting in Excel or CSV formats efficiently. With an expressive query language and document manipulation framework well integrated into the protocol, clinicians can apply such transformations to data and mold the various facets of data into the custom domain-specific model suitable to their needs.
An example of Tags used in current studies is the manual coding and entry of pencil-and-paper tests taken by participants during their first and last in-person visit. Once attached to the participant data, it can be accessed, updated, or deleted through the protocol in the same way as active or passive data from the app. This data integration allows for effective coercion and harmonization of external data sources, and therefore integrated analytics, displaying visualizations, and further research goals. An example of Automations is the download and storage of remote resources from isolated or foreign remote servers, after which different Automations can process and transform the data into mindLAMP-supported schema types. This allows multiple systems such as Apple HealthKit and Google Fit to integrate with the system and be seen by a researcher or data scientist as if it were a single unified platform.
Considering a hypothetical scenario integrating these framework components together, detailed in Figure 5, a machine learning algorithm is used by a clinician to better understand the baseline of a patient as compared to other patients with the same illness and to patients without the same illness. To automatically tune the algorithm, incoming events for each patient would be fed through an automation applet, then producing a set of outcome metrics. Supposing the algorithm reports feedback visually with an alert placed in the dashboard for the clinician to see, a new tag containing the alert is created with a pre-negotiated name known to both the algorithm and the dashboard. When the dashboard sees the tag attached to the patient, it shows the clinician along with further actions suggested by the algorithm. If the algorithm has determined that the patient is in a state of high anxiety, it may suggest the clinician to activate a soothing breathing exercise intervention. This Activity would have been configured before-hand and available to the patient, but if the clinician decides to accept the suggestion, a notification is sent to the patient that will open the Activity. The dashboard visualizations shown to both the clinician and the patient in this case would be recomputed and automatically adjusted to show data both pre and post intervention deployment. The clinician would have access to feedback data on whether the deployment of the intervention was successful or not.
Furthermore, another automation applet would in this case be configured to generate weekly reports for the patient of progress over previous weeks, highlighting whether interventions taken by the patient were successful, such as walking more often or sleeping more often. These reports are then emailed or physically mailed to the patient before their next appointment at the clinic. With automated medication tracking from auxiliary services such as Apple’s HealthKit, a full mental health profile can be managed by the clinician within one session with the patient.
Conclusions
As mobile computing and technology continue to develop rapidly, a careful standards-oriented approach must be taken to ensure the field of psychiatry is ready and able to harness it both in its current incarnation as well as embrace its future potential and possibilities. It is crucial to accept existing well-accepted standards and extend the ecosystem of tools, libraries, applications, and products built around them. We identified an unmet need in such a standard, FHIR, taking the opportunity to design a cooperative and coexisting protocol as well as develop a corresponding reference implementation. Our proof-of-concept platform, mindLAMP, is currently being used in clinical research studies across medical domains including psychiatry, anesthesiology, neurology. By architecting this standard and platform with current and well-accepted medical/psychiatric, computer engineering, and biostatistical best practices in mind, and sharing such in the public domain, we invite others to contribute, expand, and explore this intersection of digital health, data standards, and mobile healthcare.
Supplement
Supplement I Comparison of open standards
The following diagram details our research into HTTP-based transmission formats: (I) Remote Procedural Calls, (II) Resource-oriented RPC, and (III) RESTful. The model chosen in our reference implementation is indicated with a bubble.
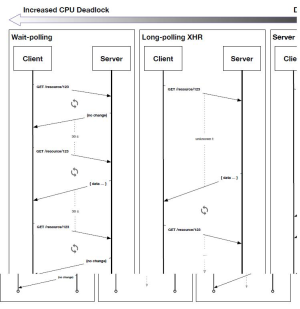
The following diagram details our research into various HTTP-based event stream delivery methods: (I) Wait-polling, (II) Long-polling/XHR, and (III) Server Push Notifications. The model chosen in our reference implementation is indicated with a bubble.
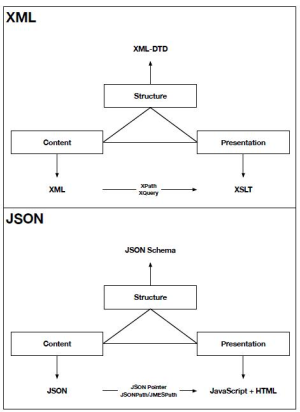
The following diagram details the differences between the XML and JSON ecosystems; additionally, XML typically is used with a SOAP transmission model where JSON is used with a REST transmission model.
Supplement II Automations in-depth
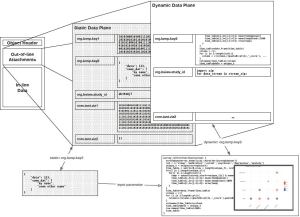
Automations and Tags are implemented as multiple planes existing on the same data format; the use of an alternation data type extension marks a Tag as an Automation to be handled differently. The visual layering in the diagram shown below corresponds to the method by which different Tags and Automations relate to one-another.
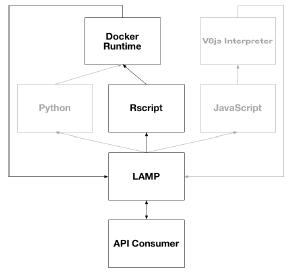
The following diagram shows the available script runtime execution pathways available for Automation applets in the current reference implementation.
The following is an example of using a JSONPath transformation to re-model data into a domain-specific schema for visualizing events in a timeline shown in a dashboard.
Acknowledgments
Funding: J Torous is supported by a career development award from the NIMH: K23MH116130.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Onnela JP, Rauch SL. Harnessing Smartphone-Based Digital Phenotyping to Enhance Behavioral and Mental Health. Neuropsychopharmacology 2016;41:1691-6. [Crossref] [PubMed]
- Insel TR. Digital Phenotyping: Technology for a New Science of Behavior. JAMA 2017;318:1215-6. [Crossref] [PubMed]
- Hsin H, Torous J, Roberts L. An Adjuvant Role for Mobile Health in Psychiatry. JAMA Psychiatry 2016;73:103-4. [Crossref] [PubMed]
- HL7. 2017. Welcome to FHIR. Retrieved Dec 14, 2018. Available online: https://www.hl7.org/fhir/
- VA announces new Veterans Health Application Programming Interface. Dec. 2018. Available online: https://www.va.gov/opa/pressrel/pressrelease.cfm?id=5158
- Bloch J. How to design a good API and why it matters. Paper presented at the Companion to the 21st ACM SIGPLAN Symposium on Object-Oriented Programming Systems, Languages, and Applications, 2006:506-7.
- Ochian A, Suciu G, Fratu O, et al. An overview of cloud middleware services for interconnection of healthcare platforms. Paper presented at the 2014 10th International Conference on Communications (COMM), 2014:1-4. doi:. [Crossref]
- Shah SP, Huang Y, Xu T, et al. Atlas - a data warehouse for integrative bioinformatics. BMC Bioinformatics 2005;6:34. [Crossref] [PubMed]
- Bray T. The javascript object notation (json) data interchange format. Internet Engineering Task Force (IETF). 2014.
- Fielding RT. Architectural Styles and the Design of Network-Based Software Architecture. Representational state transfer, 2000:76-85.
- Bryan P, Zyp K, Nottingham M. JavaScript object notation (JSON) pointer. Retrieved Dec 14, 2018. Available online: https://tools.ietf.org/html/rfc6901
- Boussadi A, Zapletal E. A Fast Healthcare Interoperability Resources (FHIR) layer implemented over i2b2. BMC Med Inform Decis Mak 2017;17:120. [Crossref] [PubMed]
- Wagholikar KB, Mandel JC, Klann JG, et al. SMART-on-FHIR implemented over i2b2. J Am Med Inform Assoc 2017;24:398-402. [PubMed]
- Patias I, Georgiev V. Mobile medical applications: From cloud-oriented to cloud ready. 2017. MCIS 2017 Proceedings 6.
- Challis RJ, Kumar S, Stevens L, et al. GenomeHubs: simple containerized setup of a custom Ensembl database and web server for any species. Database (Oxford) 2017. [Crossref] [PubMed]
- Hung LH, Kristiyanto D, Lee SB, et al. GUIdock: Using Docker Containers with a Common Graphics User Interface to Address the Reproducibility of Research. PLoS One 2016;11:e0152686. [Crossref] [PubMed]
- Ríos J, Karlsson J, Trelles O. Magallanes: a web services discovery and automatic workflow composition tool. BMC Bioinformatics 2009;10:334. [Crossref] [PubMed]
Cite this article as: Vaidyam A, Halamka J, Torous J. Actionable digital phenotyping: a framework for the delivery of just-in-time and longitudinal interventions in clinical healthcare. mHealth 2019;5:25.