
Approaches for text mining of mHealth literature
As the scientific literature search is becoming increasingly difficult to perform manually, as they do in the other areas of scientific discovery and improvement, computers are helping us automate this task too. mHealth is a relatively young research domain with an accelerated rate of scientific literature productivity, especially since mid-2010s (1). In 2017, we captured the time dependency of terminology, clusters of mHealth literature, and their trend analyses using text mining techniques (2). In 2019, Hyejin Park and Min Sook Park have followed-up with a paper that used text mining techniques to specifically study medical conditions, interventions, and study populations and medical conditions’ relationships with categories of interventions and study populations (3). In this paper, we discuss the methods and results of the two studies and how future work can improve use of text mining to capture trends in mHealth research in light of our experiences, other literature that used text mining for surveying the literature, and the evolution of text mining methods for literature review.
One of the initial steps of text mining of the literature is the search of databases to identify as many of the relevant publications as possible (establishing the corpus). This is an important step for any literature survey study; however, especially important for text mining studies, compared to manual review of the literature, because text mining methods are more accurate when there are larger data. When this work is done for a relatively younger field, where there are fewer years with research dissemination, and a field that is not strictly confined in one discipline, the impact of searching a higher number of databases is more significant in the interpretation of the results. mHealth is both a relatively younger research field and an interdisciplinary one. Furthermore, scientific literature databases may be explicitly or implicitly biased for one discipline over another, and their coverage of journals is not all-inclusive (4). Therefore, although databases such as Web of Science and Scopus are considered more interdisciplinary than databases such as PubMed and IEEE Xplore, incorporating articles from multiple databases vs. using a single interdisciplinary database would significantly impact the interpretation of results (5,6). Of course, the inclusion of articles from multiple databases introduces the problem of retrieving duplicate citations; however, most citation management tools help deal with the duplicate records issue relatively easily. In cases where the features of these tools are not sufficient, a manual duplication removal method was described in our earlier work (2). In terms of database searches, the use of search terms and the inclusion of publication types are other important considerations. We feel that providing details of the determination of search terms in a text mining publication is important for reproducibility and clarity of the process authors go through establishing the final search string. For example, inclusion of some conference publications should be considered in mHealth literature survey because disciplines like computer science that are relevant for mHealth research put a relatively much higher value on conference publications compared to other disciplines. Similar to a detailed explanation of the searching process for establishing the corpus, explaining the process of curating the term list through tokenizing, phrasing, and terming in detail is also important for the purposes of reproducibility. Finally, publishing the list of articles and the final corpus used for analyses to accompany the manuscript would improve the transparency and reproducibility of text-mining research.
Because text mining is not a field where implementations of methods are completely standardized, text mining research dissemination requires a good understanding and description of the software tool, the text mining method(s), and how the software tool implements the method(s). Text mining methods in literature review have evolved over the last few decades. Earlier methods included clustering using the document-term matrix (DTM) in high dimensional space with frequency numbers and in reduced space with singular value decomposition (SVD) outcomes. Then came latent semantic analysis (LSA) and latent dirichlet allocation (LDA) (7,8). The later trends include a neural network-based algorithm, word2vec, to produce word embeddings (9), and a deep learning technique long short-term memory (LSTM), which was introduced based on an artificial recurrent neural network (RNN) architecture (10) and applied to text mining of literature in numerous examples (11-14).
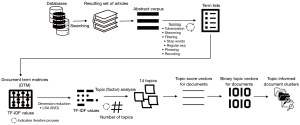
Hyejin Park and Min Sook Park claimed that our study used most frequently surfaced terms using clusters, which were topic analysis (TA) informed document clusters, and that our method was “limited in determining details of relationships among terms and producing in-depth understanding” (3). In our study, we used a combination of two methods, namely document clustering (DC) using latent class analysis (LCA) and TA using LSA, the process of which is described in Figure 1. These analyses are performed on the DTM. For each cell of DTM, instead of simple term frequency (TF) that indicates the number of times a term is used in a given document, we used TF into inverse document frequency (TF-IDF) that indicates how important a term is to a document in a corpus, in order to avoid potential bias caused by high term frequencies in a document (IDF is computed as the logarithm of the number of the documents in the corpus divided by the number of documents where the specific term appears, and TF-IDF is multiplication of TF by IDF). LCA generates hierarchical clusters of documents. Since the resulting term-sets for those clusters were not sufficiently distinct, we decided to include TA to inform DC. LSA is equivalent to performing principal components analysis (PCA), centered around computing a partial SVD. TA in our study was basically equivalent to a varimax rotation on the partial SVD of the DTM. Normally, TA is independent of document clusters created by LCA; however, we captured document topic vectors and assigned each document to a topic based on their topic scores, allowing us to create document clusters informed by TA. In other words, our study performed a TA that not only considered relationships among the terms, but also considered closeness of those relationships by performing TA. Hence, we disagree with Hyejin Park and Min Sook Park’s characterization of our study as limited in determining details of relationships among terms.
Hyejin Park and Min Sook Park used co-occurrence networks that only showed the pattern of paired terms appearing together, rather than a full TA that considers the entire terms set and provides a set of representative terms for the topic. They limited terms to be included in the co-occurrence network analysis to top ten terms that represent each of the three manually assigned term sets, namely medical conditions, interventions, and study populations. In a sense, they manually performed a version of TA by having each of the two researchers assign terms into one of these three sets. Combining terms into subjectively defined topics is a good idea when we know that those topics are important for the field. Although it may be obvious, Hyejin Park and Min Sook Park did not explain how they decided to study medical conditions, intervention types, and study populations. They could have applied a process of reviewing the initial terms list and recognizing these categories in the list or deciding on these categories beforehand, assuming most mHealth solutions would have these three components. Future studies may do a combination of TA for the machine to tell us what topics it identifies and researchers determining what categories are interesting for the field, also explaining the reasoning for this subjective determination. When categorizations of terms are deemed necessary, use of terminologies, ontologies, and taxonomies for this purpose, like Hyejin Park and Min Sook Park did by using MeSH Subject headings for medical conditions and intervention taxonomy (ITAX) for interventions, would be ideal. Future studies could explore how to automate this term categorization by using a modified TA method where TA is informed by prior knowledge of an existing terminology (hierarchical or otherwise), instead of solely depending on PCA of independent terms.
In conclusion, we believe both studies have contributed to our understanding of the mHealth literature from different perspectives utilizing some of the common methods, as well as some distinct ones. We also believe that future studies could utilize a combination of these methods and more recent methods such as LSTM, and increase the type and volume of results and interpretations. Furthermore, future studies could perform post-categorization correlation (15) or correspondence (16) analyses, examples of which are implemented in our works performing text mining of the literature in other domains. Others have also published examples that used full-text of articles as opposed to titles and abstracts only (17), utilized heatmaps to visualize topic clusters and co-occurrence networks to visualize high-loading terms (18), and other network visualizations to demonstrate topic similarities (19).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, mHealth. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://mHealth.amegroups.com/article/view/10.21037/mHealth-22-1/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cameron JD, Ramaprasad A, Syn T. An ontology of and roadmap for mHealth research. Int J Med Inform 2017;100:16-25. [Crossref] [PubMed]
- Ozaydin B, Zengul F, Oner N, et al. Text-mining analysis of mHealth research. mHealth 2017;3:53. [Crossref] [PubMed]
- Park H, Park MS. Capturing the trend of mHealth research using text mining. mHealth 2019;5:48. [Crossref] [PubMed]
- Mongeon P, Paul-Hus A. The journal coverage of web of science and scopus: a comparative analysis. Scientometrics 2016;106:213-28. [Crossref]
- Cooper C, Booth A, Varley-Campbell J, et al. Defining the process to literature searching in systematic reviews: a literature review of guidance and supporting studies. BMC Med Res Methodol 2018;18:85. [Crossref] [PubMed]
- Gough D, Oliver S, Thomas J. editors. An introduction to systematic reviews. 2nd edition. ed. Los Angeles: SAGE, 2017.
- Kim YM, Delen D. Medical informatics research trend analysis: a text mining approach. Health Informatics J 2018;24:432-52. [Crossref] [PubMed]
- Miner G, Elder J, Fast A, et al. editors. Practical text mining and statistical analysis for non-structured text data applications. 1st Edition. Amsterdam: Elsevier, 2012.
-
Mikolov T Chen K Corrado G Efficient estimation of word representations in vector space. arXiv:13013781.2013 . - Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997;9:1735-80. [Crossref] [PubMed]
- Gajendran S, D M, Sugumaran V. Character level and word level embedding with bidirectional LSTM - Dynamic recurrent neural network for biomedical named entity recognition from literature. J Biomed Inform 2020;112:103609. [Crossref] [PubMed]
- Park Y, Lee J, Moon H, et al. Discovering microbe-disease associations from the literature using a hierarchical long short-term memory network and an ensemble parser model. Sci Rep 2021;11:4490. [Crossref] [PubMed]
- van der Vegt AH, Zuccon G, Koopman B. Learning inter-sentence, disorder-centric, biomedical relationships from medical literature. AMIA Annu Symp Proc 2020;2019:1216-25. [PubMed]
- Yu Y, Si X, Hu C, et al. A review of recurrent neural networks: LSTM cells and network architectures. Neural Comput 2019;31:1235-70. [Crossref] [PubMed]
- Zengul FD, Zengul AG, Mugavero MJ, et al. A critical analysis of COVID-19 research literature: text mining approach. Intell Based Med 2021;5:100036. [Crossref] [PubMed]
- Zengul AG, Zengul FD, Ozaydin B, et al. Identifying research themes and trends in the top 20 cancer journals through textual analysis. J Cancer Policy 2021;30:100313. [Crossref]
- Abdelrahman MM, Zhan S, Miller C, et al. Data science for building energy efficiency: a comprehensive text-mining driven review of scientific literature. Energy and Buildings 2021;242:110885. [Crossref]
- Romero-Silva R, De Leeuw S. Learning from the past to shape the future: a comprehensive text mining analysis of OR/MS reviews. Omega 2021;100:102388. [Crossref]
- Cheng X, Cao Q, Liao SS. An overview of literature on COVID-19, MERS and SARS: using text mining and latent Dirichlet allocation. Journal of Information Science 2020;1-17. [Crossref]
Cite this article as: Ozaydin B, Zengul F, Oner N, Delen D. Approaches for text mining of mHealth literature. mHealth 2022;8:11.


